Data Reduction
Reduction is the process of converting Raw data which is a list of time stamped events and the associated metadata to a file in units proportional to S(Q,ω). In this process various intermediate files may be produced. The main file formats are nxs and nxspe both of which are hdf5 and can be browsed with hdf5 viewing tools. Reduction is handled by the Mantid routines.
File formats
Raw files
The raw data is stored in HDF5 files formatted according to the NeXus event file standard.
In this file each neutron event is recorded as a pixel id and a time stamp.
These files also include logs and the instrument information.
They are located at /SNS//IPTS-XXXX/nexus/_YYYY.nxs.h5 where XXXX is the IPTS number and YYYY is the run number and are catalogued in OnCat.
Reduced files
-
nxs files
This is simply a NeXus file which can encompass many types of data.
-
nxspe files:
This format is a histogram format the main data structure contains counts in bins of Energy transfer and pixel id. The mapping of pixel id to scattering angle is also contained in the file. Mantid, DAVE, and Horace are all designed to work with the file format.
-
spe/par files:
These files are ASCII and are legacy and should not be created.
Nevertheless they are identified here for reference purposes. The nxspe format contains all the information of the spe and par files and is much more efficient for storage. The most used visualization packages (Mantid, DAVE, and Horace) all can read the nxspe format.
Autoreduction
Data is usually autoreduced. When a runs finishes a python script located at
/SNS/ARCS/shared/autoreduce/reduce_ARCS.py is executed and provides files for use in Visualization and analysis. Your local contact ensures that it is setup and can modify it if necessary. Usually the reason to modify it would be for adjusting energy bins. The results from the autoreduction procedure are saved in /SNS/ARCS/IPTS-XXXX/shared/autoreduce. Both nxspe and nxs files are saved in this directory.
Also the powder averaged data is shown on monitor.sns.gov. See the web monitor pages for more information
Manual Reduction
A good way to start a manual reduction is to start from the Autoreduction script. If you copy it to your home directory or the shared directory of your IPTS, you can modify it and run it from the command line.
Masking Detectors
Sometimes not all the detectors are useable (e.g. they have a known spurious feature or they are blocked by sample environment) and thus the user would like to remove them from the reduction process.
There are several tools to generate a mask.
All of them start with identifying the detectors you want to mask.
To do this:
- Open MantidWorkbench
- Load the raw datafile (see the above description of where they are located) you wish to mask.
-
Open instrument view by right clicking on the workspace and selecting “Show Instrument”
 and the instrument view will appear
and the instrument view will appear
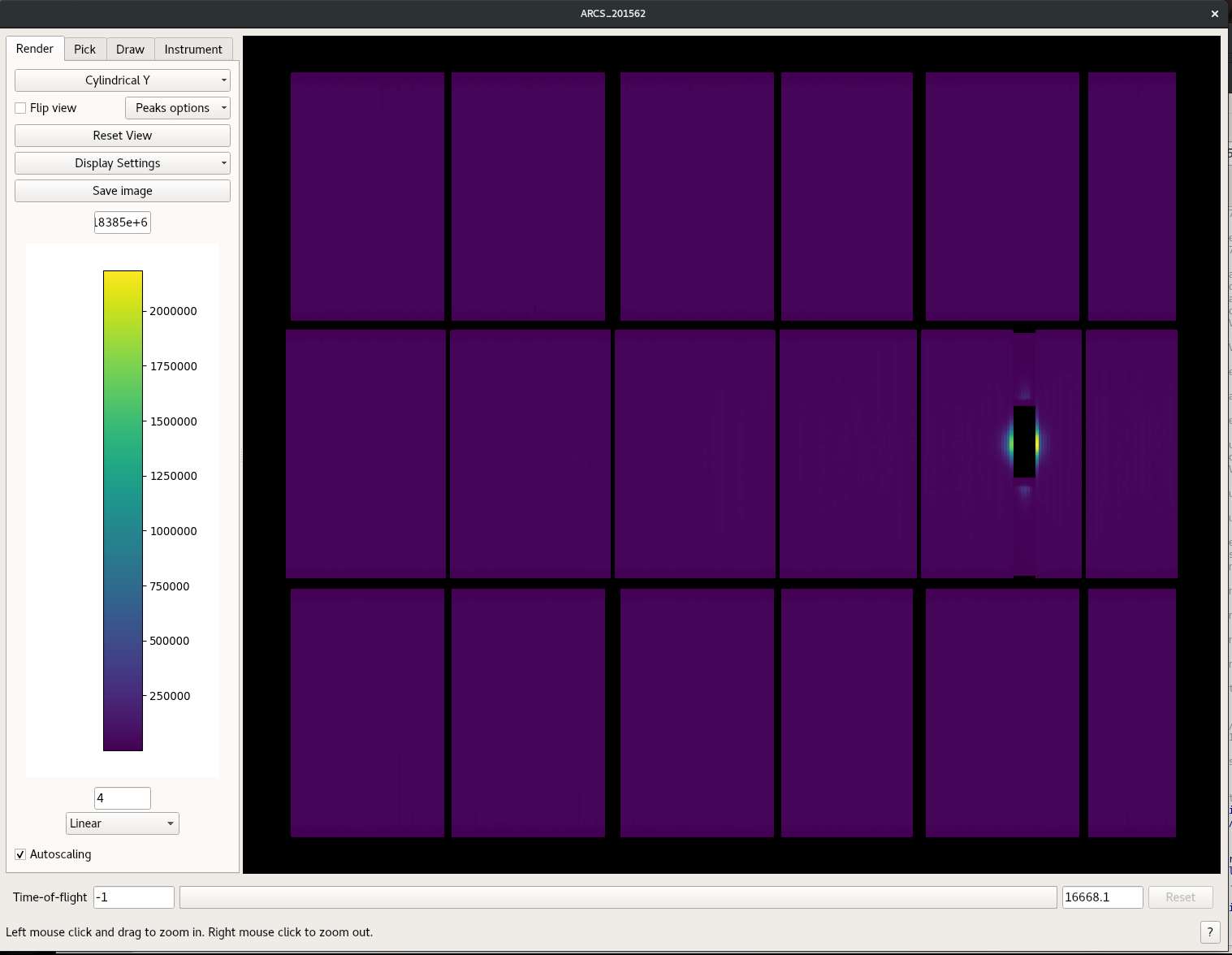
- generate the Mask in one of the 2 ways provided below
- Save a masked workspace
- Load that mask workspace and use the Mask Detectors algorithm on all subsequent workspaces you wish to mask
There are two ways to generate the mask
First you can determine the bank tube and pixel identifier and use the MaskBTP algorithm.
In instrument view you select the pick tool and hover over the pixel you want to mask.
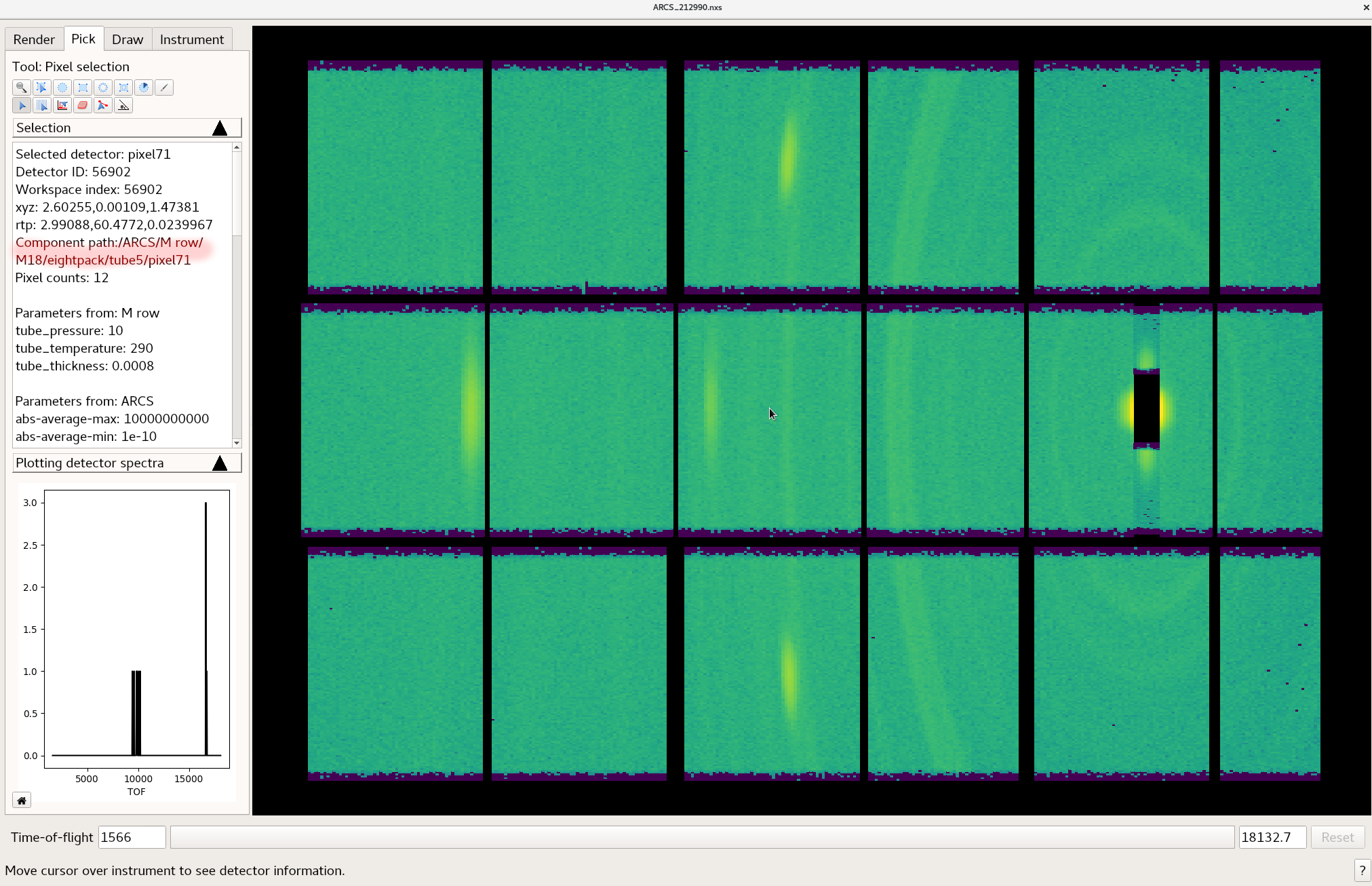 An identifier for the bank, tube, and pixel
will be provided as is indicated by the highlighted area in the figure.
The bank number is of a different convention to that used in the MaskBTP algorithm.
Use the table in detector description
An identifier for the bank, tube, and pixel
will be provided as is indicated by the highlighted area in the figure.
The bank number is of a different convention to that used in the MaskBTP algorithm.
Use the table in detector description
Second you can mask right in the instrument window.
- Select the Draw tab as shown in the following image
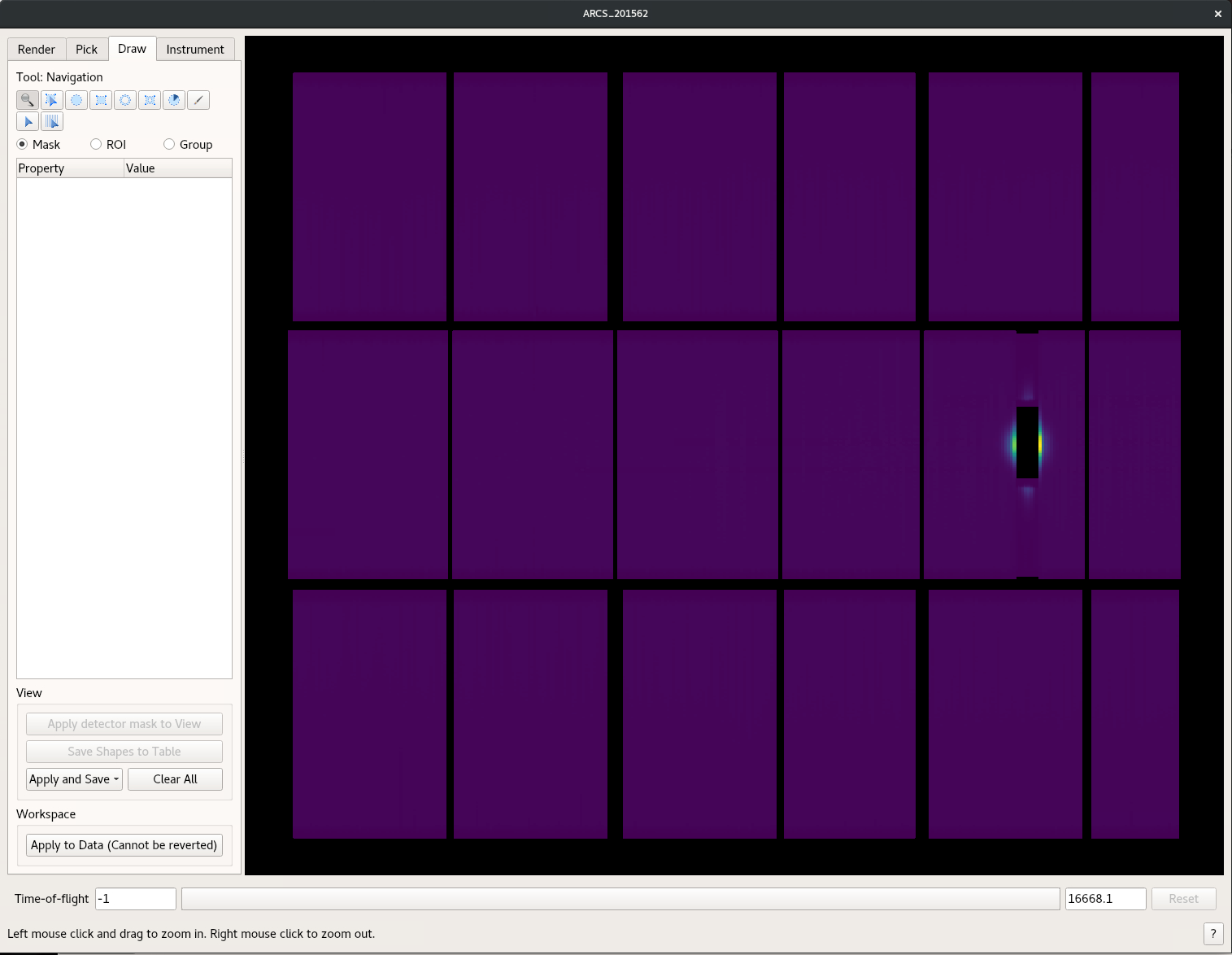
- Draw what ever shape you like the grey box will then be masked. You can draw multiple masks
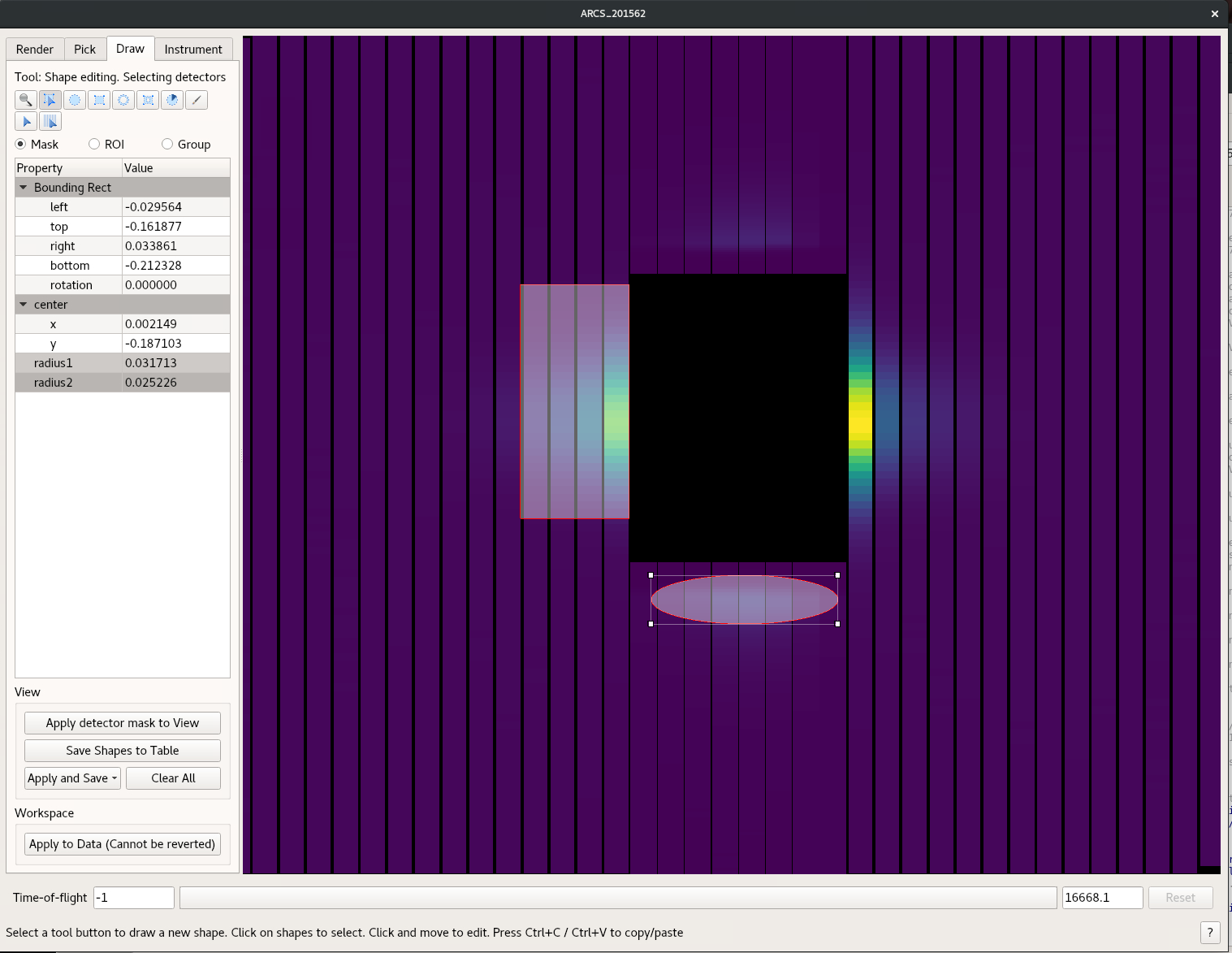
- Once you are done use the Apply and Save button and chose the “As Detector Mask to Workspace”
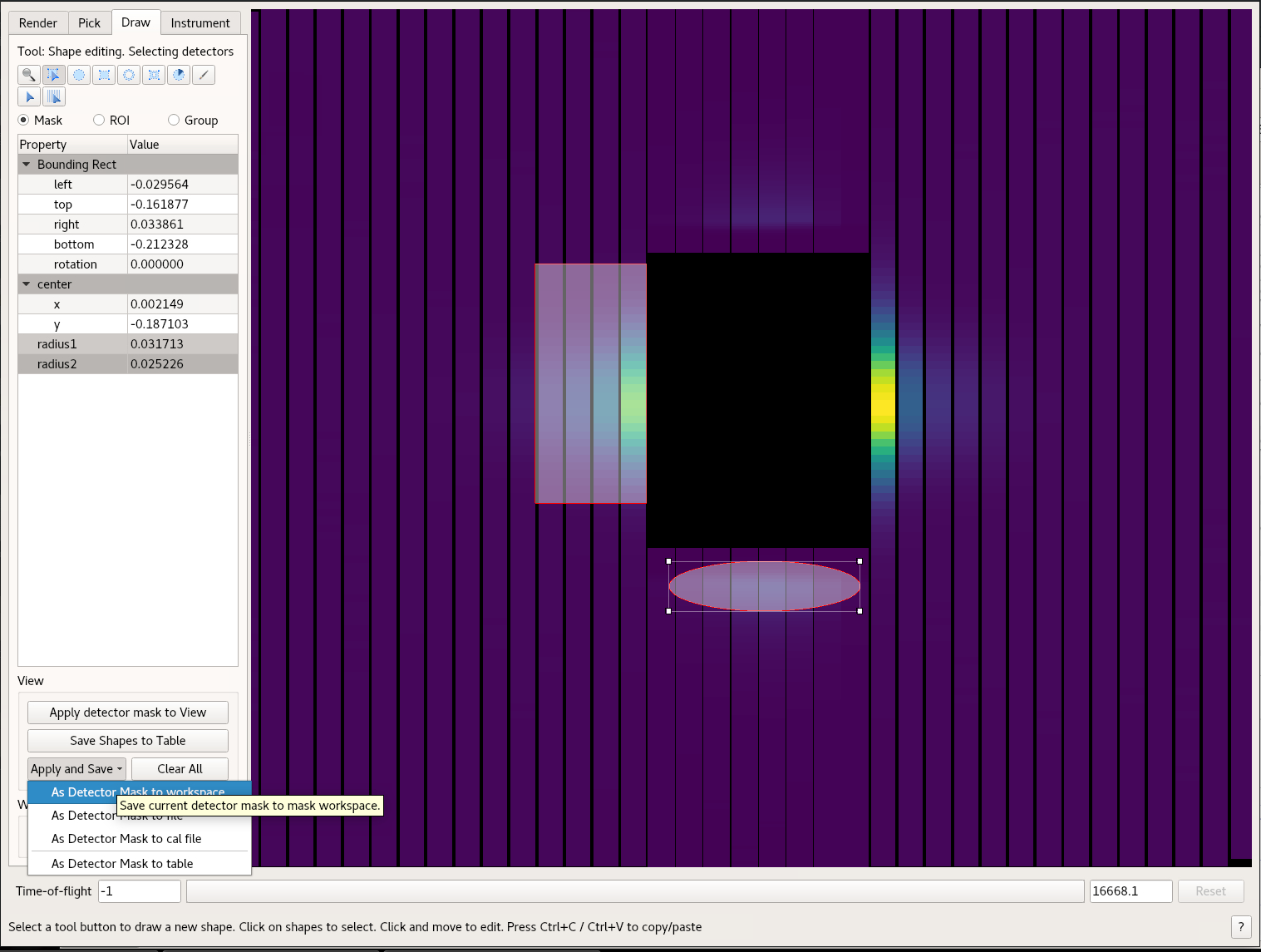
In the workspace window in Mantid Workbench, you will see MaskedWorkspace. This is the mask. Use the MaskDetectors algorithm to apply this mask to another workspace. Use the MaskedWorkspace option.
More specifically we usually mask the V workspace so the mask can be used in further reductions. To do this
- Load the Vanadium Workspace
- Open the MaskDetectors Algorithm
- In the MaskDetectors Algorithm
- Put the name of the Vanadium workspace in the InputWorkspace box
- Select your masked work space from the MaskedWorkspace pull down
- Press Run
- Once the Algorithm is done Press Save and Save as nexus. Be sure to save it to a different name
Determining a Transmission function for the the Radial Collimator
The transmission through the radial collimator varies as a function of scattering angle and Ef.
This transmission function is flat above in plane scattering angles of 40o ( M. B. Stone et al. EPJ Conf. 83, 03014 ). So for most phonon studies it is best to just ignore the data at low scattering angles.
If this region is critical one needs to calculate a transmission function.
For Ef < 100 meV, there is no energy dependence on the transmission function. Therefore the following procedure will calculate this function. The data needed for this procedure is two monochromatic V runs at the same Ei value as the data to be corrected.
The first V run should be with the radial collimator in and the second one with the radial collimator out.
With these two data sets, the steps are as follows.
- Load both data sets.
- Rebin each data set to be 1 bin between 100 μs and the full length of the frame.
- Normalize each data set by current.
- Divide the with collimator data set by the without collimator data set.
- Then in the reduction multiply the V normalization data set by the just calculated transmission function to correct the V Normalization.
Absolute Normalization
The reduction process takes the collected data to a function proportional to dσ2/dΩdω. Determining the proportionality constant is often not necessary to determine the experimental result. Furthermore this constant usually has the largest uncertainty. But there are cases where an absolute measurement is required. So methods to calculate it are described below.
Calculating the constant is highly sample dependent so three methods are described. These methods are summarized in G. Xu, Z. Xu, and J. Tranquada, Review of Scientific Instruments 84, 083906 (2013)
Absolute Normalization with V
This is the most common method.
For this measurement a monochromatic V run is needed.
The instrument staff do several common incident energies at the beginning of a run cycle so they may have this measurement for you.
If your incident energy has not been preformed, you will need to do this.
For this measurement Your instrument staff will help you set it up:
- The V is smaller than the beam.
- The V mass is known
- The V was heat treated to remove any H left from the casting process.
- A room temperature stick is used to remove scattering from sample environment.
This method assumes the sample illumination is the same as the V illumination.
For instruments with no guide or for measurements where the incident energy is above where the guide plays a significant role in increasing the flux on sample, this assumption is completely valid.
Outside these limits a correction should be applied.
Absolute Normalization with a Phonon
Absolute Normalization from Sample incoherent scattering
Event Filtering
As the raw data is in events it can be filtered and then each subset processed. This usually means writing a manual reduction script. Your Instrument staff can help you with this. Generally Event filtering has these steps.
- Read the events
- Do as many event based operations as possible ( ie convert to Q and ω)
- determine how the events should be filtered by using the GenerateEventsFilter algorithm.
- Filter the events using FilterEvents algorithm.
- Complete the reduction on each filtered workspace. A workspace group is very effective for this.
Reduction Details
Reduction is handled by the DGSReduction algorithm in Mantid.
The steps in Reduction are:
1) Use the Beam monitors to determine the incident energy 1) Determine the appropriate mask for the detectors. 1) Apply the mask to a white beam V file. 1) Apply an optional transmission function correction for Radial Collimator 1) Normalize the Detector efficiency, correct for solid angle, and mask detectors using the white beam V. 1) Normalize to proton charge. 1) Convert Time of flight to energy transfer 1) Correct for ki/kf.
HDF5 viewing Tools
Sometime it is useful to view the HDF5 file there are several tools to do this
HDFView
Is a Java based tool. It is installed on the analysis machines. At the command line type hdfview.
Nexpy
Nexpy Is a Python tool for working with NeXus files. Some users find this quite useful. It is not available on the analysis machines but IT can be installed ON your local machine using conda python distribution.
Others
For low level manipulation of hdf5 files we recommend using the h5py library for Python.
Matlab, IgorPro, IDL, and Origin all have similar abilities to work with HDF5 files.
